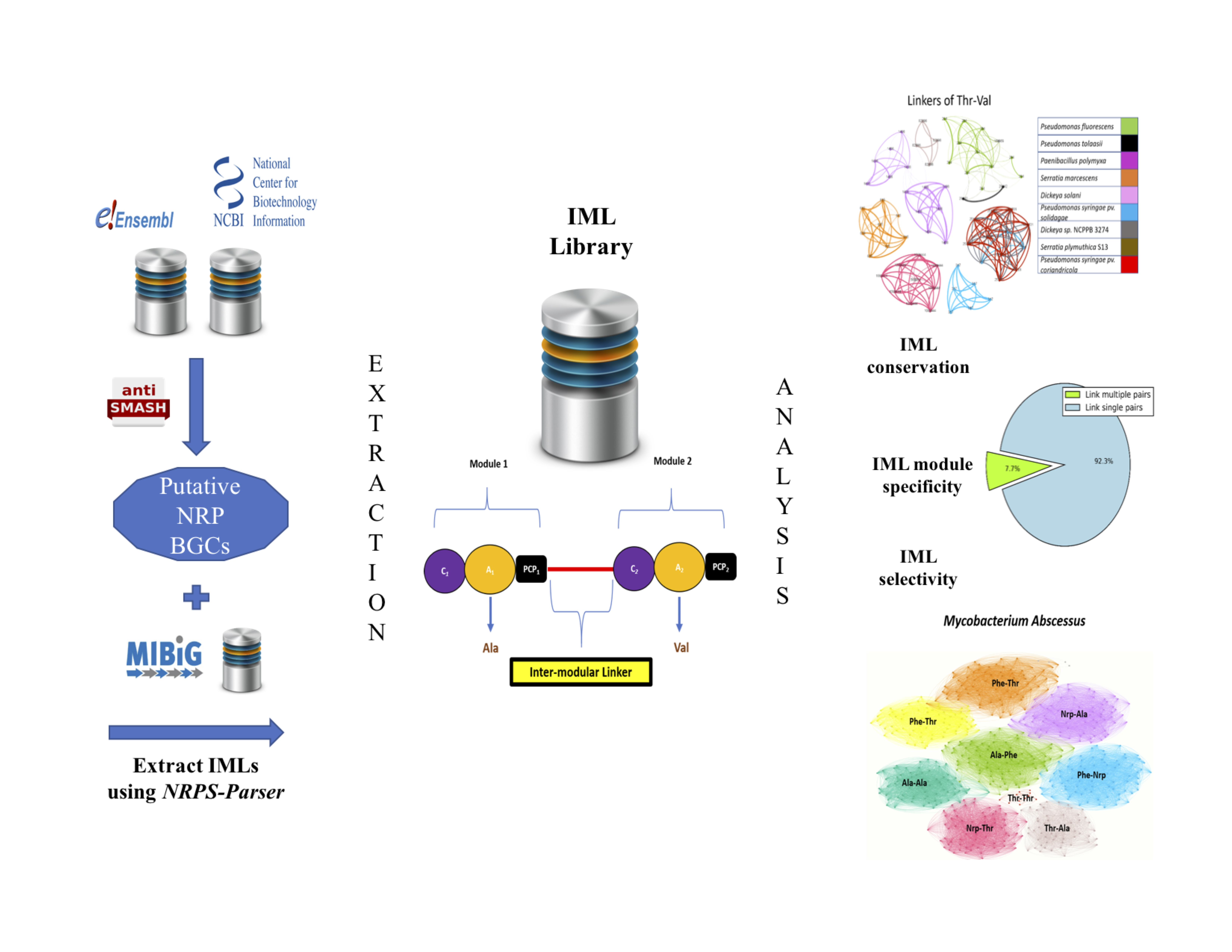
General Workflow
How selective IMLs are toward pairs of modules
-
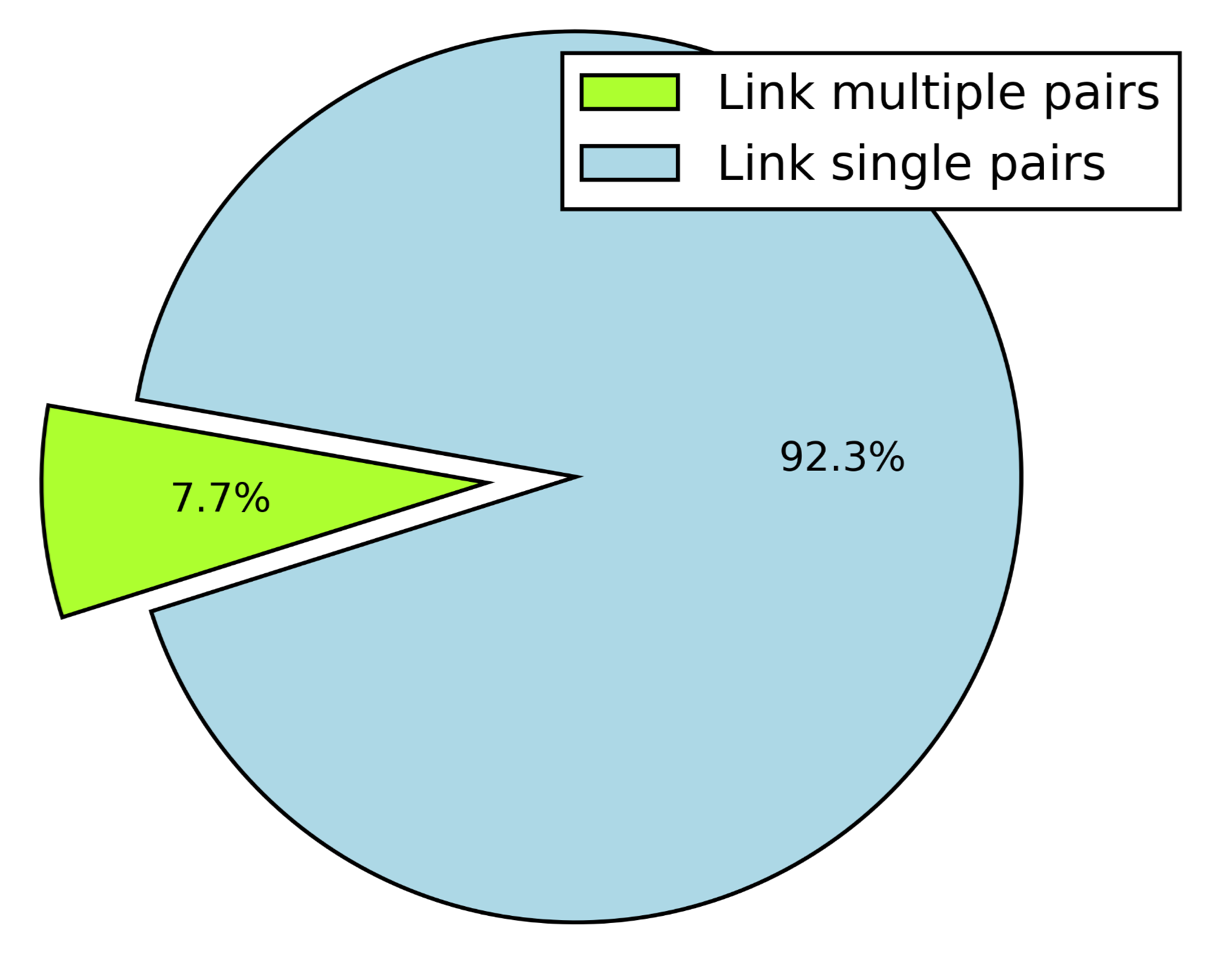
Here we are trying to explore whether linkers tend to bind multiple pairs of modules (universal linkers) or every unique linker binds a specific pair of modules. Clustering of the ~38k extracted inter-modular linkers (IMLs) using the UCLUST clustering algorithm (Edgar RC.), have shown that the data comprise of 3,916 unique linkers (centroids/clusters). 3,616 IMLs were found to be associated with only a single pair of modules, while the remaining 300 linkers tend to bind multiple distinct pairs of modules (ranging between 2 and up to 13 unique pairs) (Fig. 1).

-
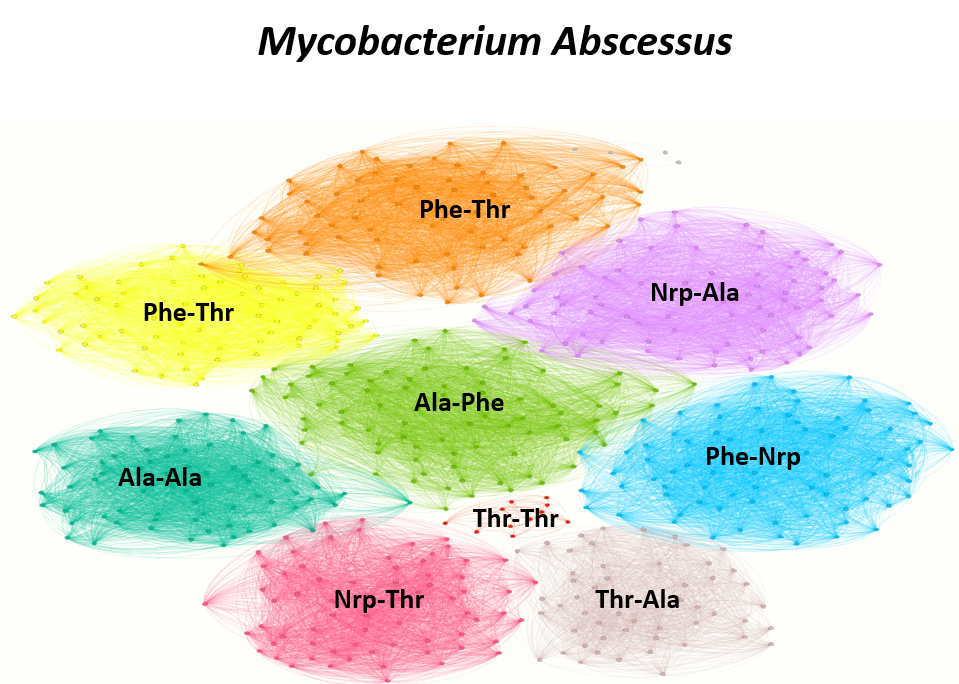
In order to establish a more rigorous case about our previous analysis, we decided to create a graph with 993 Inter-modular linkers extracted from the species Mycobacterium Abscessus. The nodes of the graph represent the linkers and edges are constructed between nodes, that share at least 80% pairwise sequence similarity. After constructing the graph, we applied Louvain community detection algorithm(Vincent D. Blondel et.al). The algorithm was able to detect 9 distinct communities. Each community consists of linkers that links specifically a single pair of modules. For simplicity purposes, a pair of modules are represented by their activated substrates (e.g. Ser-Linker-Ala --> Ser-Ala) (Fig. 2).
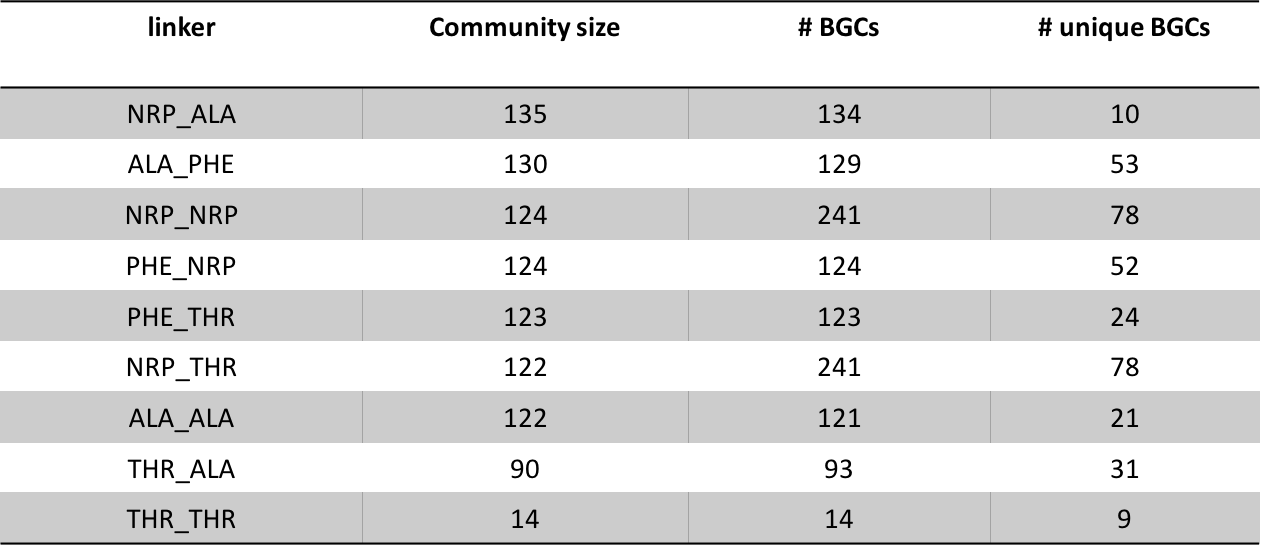
Conclusion:
From the aboved mentioned analyses we concluded, that Linkers are highly selective towards pairs of modules and they do not link them randomly.How conserved are IMLs within and across species:
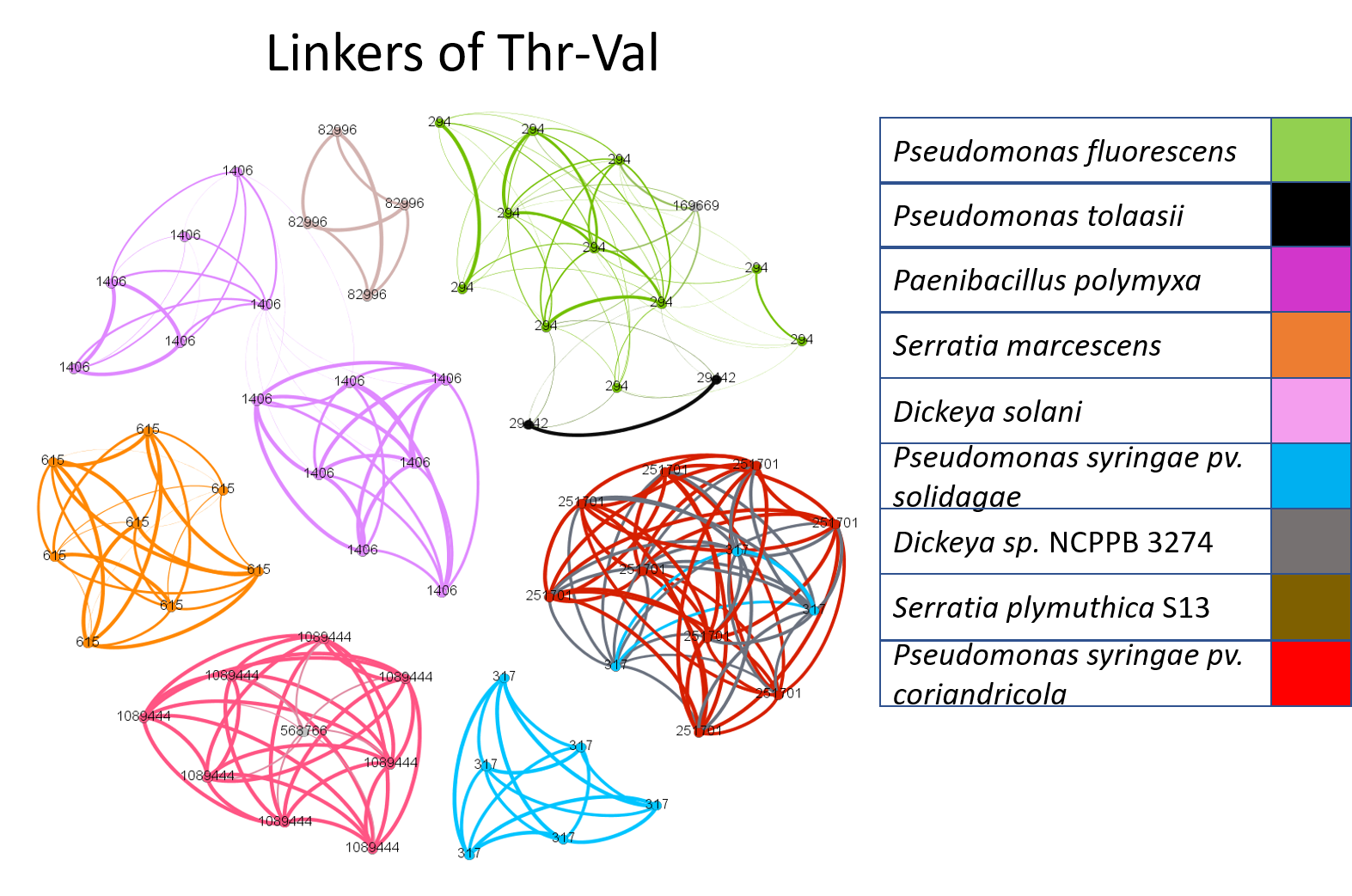
Our analysis so far indicates that IMLs are very selective towards module pairs. Here, we ask whether pairs of modules tend to be linked by the same linker regardless of the bacterial species they were obtained from. We conducted an all-by-all comparison of module pairs vs. genera (computing the degree of conservation of IMLs linking a specific module pair both within and across genera) and then built a community network to visualize the phylogenetic distributions of IMLs that link the same module pair. We then constructed a community network visualization to illustrate the phylogenetic specificity of IMLs. We took all of the Thr-Val IMLs obtained across all species and created a graph where again the nodes represent IMLs and edges are drawn only between nodes that share at least 80% sequence similarity (Figure 8). After applying the Louvain community detection algorithm to this data, the nodes were colored based on the bacterial species they were obtained from. If IMLs were globally conserved across many bacterial species, we would have expected to obtain a single large community network with multi-colored nodes. If instead IMLs were conserved within a single bacterial species, we would expect multiple distinct communities to be detected, where nodes within each community had the same color. The data indicate that the latter is the case, and that there is some phylogenetic specificity to IMLs (Fig. 3)